Results
Please cite:
if you find this tool useful in your research. Thanks!
Haplotype Analysis:
Haplotypes with frequency <0.03 are ignored.
Loci chosen for haplotype analysis: rs776746, rs2242480, rs4646437, rs1057868, rs2276707, rs227707, rs6785049, rs1523127, rs2257401, rs10211, rs12360
| Haplotype | Case(freq) | Control(freq) | Chi2 | Fisher's p | Pearson's p | OR [95% CI] |
|---|---|---|---|---|---|---|
| CCGTCAAAGTA | 18(0.095) | 18(0.096) | 0.007 | 0.999 | 0.928 | 0.969 [0.489~1.919] |
| CCGCCAAAGTA | 23(0.122) | 17(0.091) | 0.814 | 0.408 | 0.366 | 1.352 [0.7~2.612] |
| TTACTAGACCG | 14(0.074) | 10(0.053) | 0.591 | 0.53 | 0.441 | 1.386 [0.601~3.194] |
| CCGCTAGAGTA | 18(0.095) | 26(0.139) | 1.888 | 0.202 | 0.169 | 0.642 [0.341~1.211] |
| TTACCAAACCG | 8(0.042) | 4(0.021) | 1.258 | 0.381 | 0.262 | 1.98 [0.587~6.68] |
| CCGTTAGAGTA | 12(0.063) | 8(0.043) | 0.724 | 0.493 | 0.394 | 1.485 [0.594~3.71] |
| CCATCAAAGTA | 3(0.015) | 9(0.048) | 3.266 | 0.083 | 0.07 | 0.314 [0.083~1.177] |
Global result:
Total control=104, total case=107.
Global Chi2 is 8.073, Fisher's p is NA, Pearson's p is 0.232.
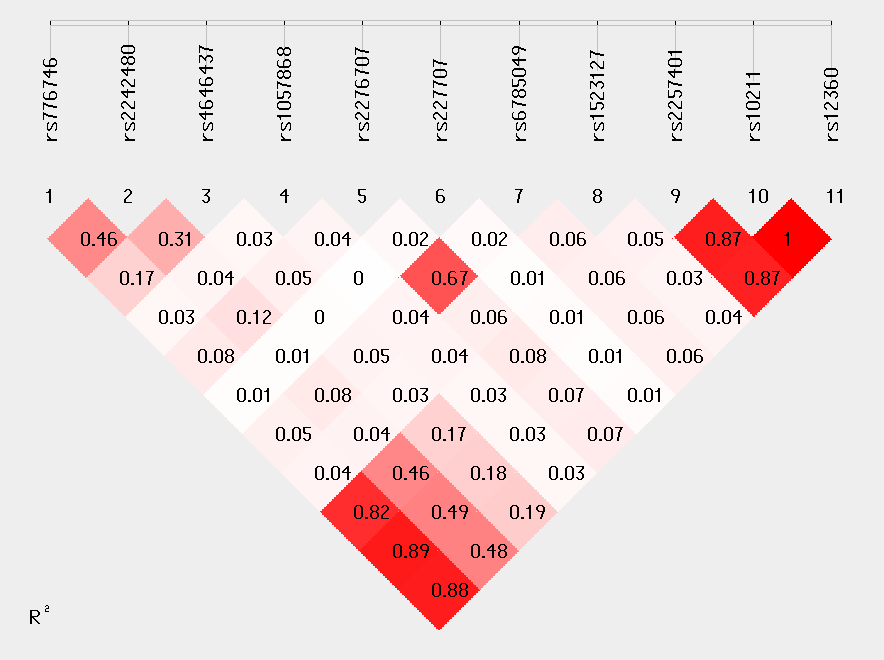
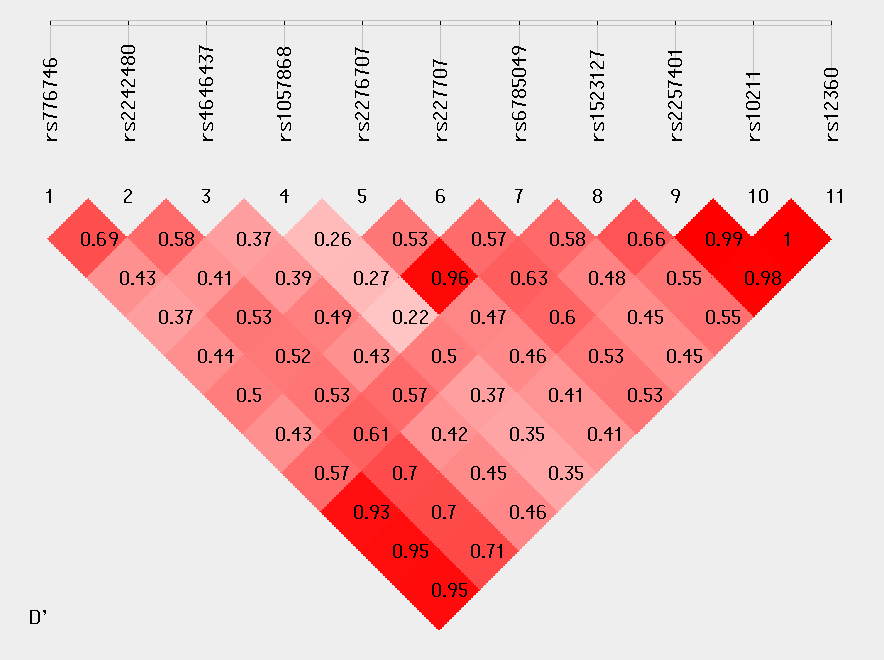
Linkage Disequilibrium Analysis:
D'
R2